VibrioNet
 Vibrio spp. are gram-negative, facultatively anaerobic, mainly halophilic and non-sporulating curved rod bacteria, typically found in the aquatic environment. The genus Vibrio comprises many species of which only 12 are known to cause disease in humans. Current knowledge is limited about the virulence determinants that are responsible for the different clinical presentations of infections with non-cholerae Vibrio spp. (diarrhea, wound infections, sepsis), such as V. parahaemolyticus or V. vulnificus. In recent years, worldwide infections with non-cholerae Vibrio spp. have increased. Global warming with elevated seawater temperatures as well as a growing global consumption and trade of seafood are amongst the reasons for this phenomenon. Vibrioses may represent an emerging but neglected zoonotic disease in Germany and other countries.
Vibrio spp. are gram-negative, facultatively anaerobic, mainly halophilic and non-sporulating curved rod bacteria, typically found in the aquatic environment. The genus Vibrio comprises many species of which only 12 are known to cause disease in humans. Current knowledge is limited about the virulence determinants that are responsible for the different clinical presentations of infections with non-cholerae Vibrio spp. (diarrhea, wound infections, sepsis), such as V. parahaemolyticus or V. vulnificus. In recent years, worldwide infections with non-cholerae Vibrio spp. have increased. Global warming with elevated seawater temperatures as well as a growing global consumption and trade of seafood are amongst the reasons for this phenomenon. Vibrioses may represent an emerging but neglected zoonotic disease in Germany and other countries.
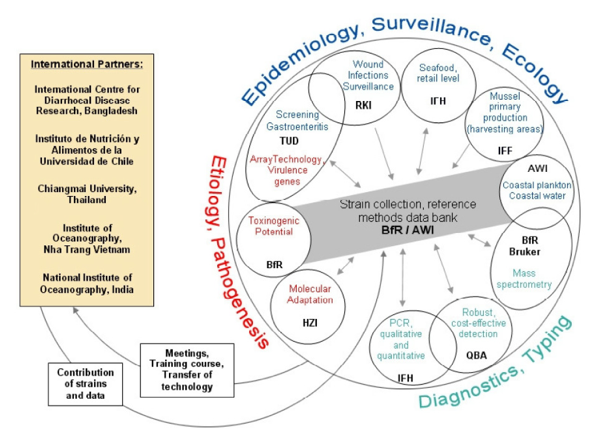 The network “VibrioNet - Climate warming and the emergence of seafood and waterborne vibrioses” consists of 7 national and 5 associated international partners.
The network “VibrioNet - Climate warming and the emergence of seafood and waterborne vibrioses” consists of 7 national and 5 associated international partners.
Our group has currently two subprojects in this network:
| 1. “Incidence of Vibrio spp. isolates in human stool specimens and evaluation of their virulence potential in in vivo model systems” | 2. “Molecular characterization of virulence determinants of pathogenic Vibrio spp. by whole genome sequencing” |
The subprojects are focused on the following aim:
|
|